pacman::p_load(seriation, dendextend, heatmaply,
tidyverse, ggridges, ggtext,
patchwork)Hands-on Exercise 5c: Heatmap for Visualising and Analysing Multivariate Data
1 Overview
Heatmaps visualise data through variations in colouring. When applied to a tabular format, heatmaps are useful for cross-examining multivariate data, through placing variables in the columns and observation (or records) in rows and colouring the cells within the table. Heatmaps are good for showing variance across multiple variables, revealing any patterns, displaying whether any variables are similar to each other, and for detecting if any correlations exist in-between them.
In this hands-on exercise, we will gain hands-on experience on using R to plot static and interactive heatmap for visualising and analysing multivariate data.
2 Getting Starting
In this hands-on exercise, the data of World Happiness 2018 report will be used. The original data set is in Microsoft Excel format. It has been extracted and saved in csv file called WHData-2018.csv.
wh <- read_csv("data/WHData-2018.csv")The output is a tibble data frame called wh, with 156 observations (rows) across 12 variables (columns).
glimpse(wh)Rows: 156
Columns: 12
$ Country <chr> "Albania", "Bosnia and Herzegovina", "B…
$ Region <chr> "Central and Eastern Europe", "Central …
$ `Happiness score` <dbl> 4.586, 5.129, 4.933, 5.321, 6.711, 5.73…
$ `Whisker-high` <dbl> 4.695, 5.224, 5.022, 5.398, 6.783, 5.81…
$ `Whisker-low` <dbl> 4.477, 5.035, 4.844, 5.244, 6.639, 5.66…
$ Dystopia <dbl> 1.462, 1.883, 1.219, 1.769, 2.494, 1.45…
$ `GDP per capita` <dbl> 0.916, 0.915, 1.054, 1.115, 1.233, 1.20…
$ `Social support` <dbl> 0.817, 1.078, 1.515, 1.161, 1.489, 1.53…
$ `Healthy life expectancy` <dbl> 0.790, 0.758, 0.712, 0.737, 0.854, 0.73…
$ `Freedom to make life choices` <dbl> 0.419, 0.280, 0.359, 0.380, 0.543, 0.55…
$ Generosity <dbl> 0.149, 0.216, 0.064, 0.120, 0.064, 0.08…
$ `Perceptions of corruption` <dbl> 0.032, 0.000, 0.009, 0.039, 0.034, 0.17…n_distinct(wh$Country)[1] 156Ultimately, we want a heatmap where the different countires are shown along one axis, the explanatory variables on another aixs, and the shading/intensity of the cells should reflect the magnitude of values.
Target Variable
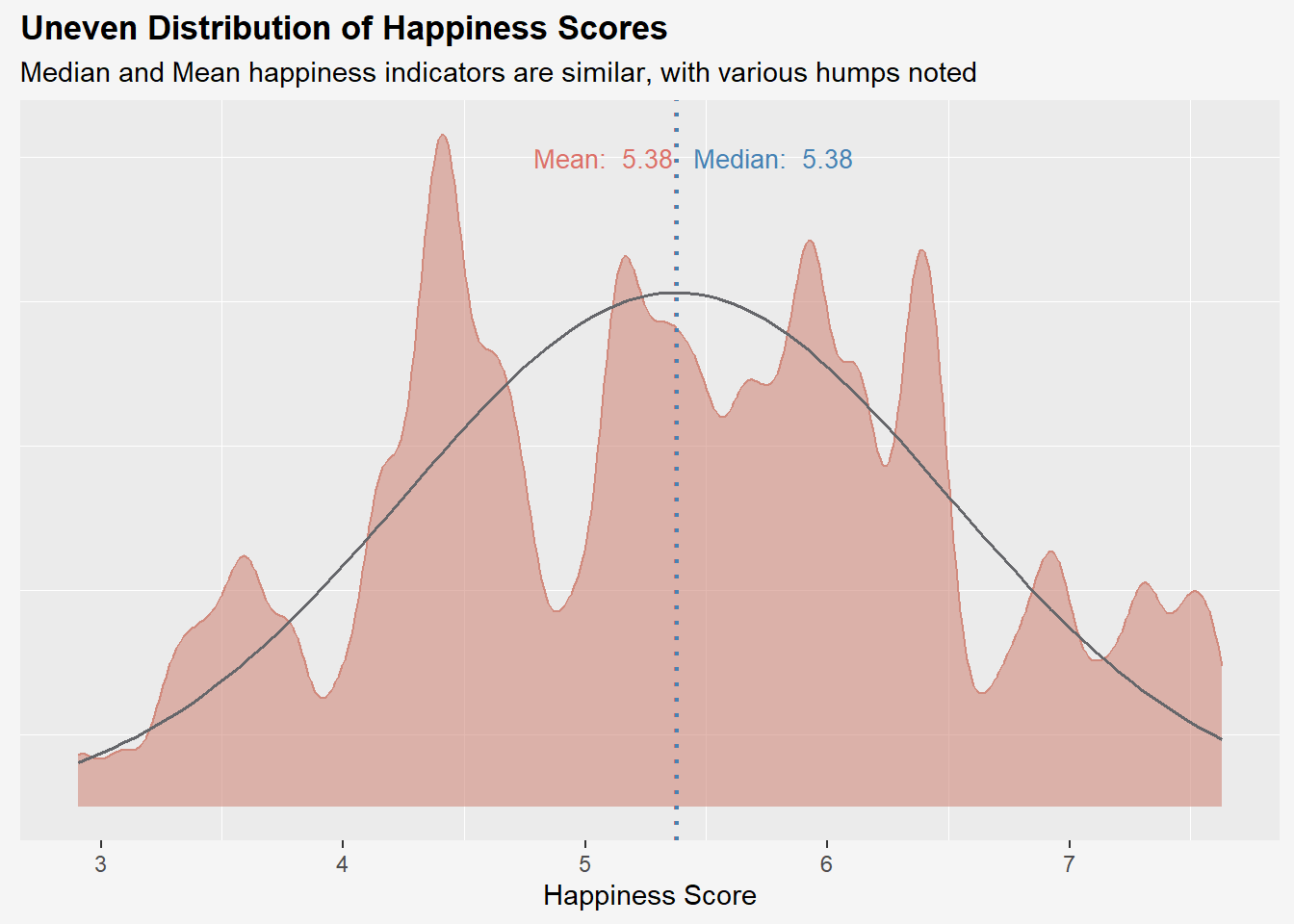
# Calculate Median and Mean values for plotting normal curve
med_hs <- median(wh$`Happiness score`)
mean_hs <- mean(wh$`Happiness score`)
std_hs <- sd(wh$`Happiness score`)
ggplot(
wh,
aes(x = `Happiness score`)
) +
# Density plot of target variable
geom_density(
color = "#d18a7d",
fill = "#d18a7d",
# Adjust bandwidth to reduce smoothness of density plot to reveal variations
adjust = .2,
alpha = .6
) +
# Plot normal curve
stat_function(
fun = dnorm,
args = list(mean = mean_hs,
sd = std_hs),
col = "#65666a",
size = .6
) +
# Adding mean line with annotation
geom_vline(aes(xintercept = mean_hs,
),
color = "#dc7068",
linetype = "dotted",
linewidth = .8,
hjust = 0.6,
size = 3) +
annotate(
"text",
x = mean_hs-0.3,
y = 0.45,
label = paste("Mean: ", round(mean_hs, 2)),
color = "#dc7068",
size = 3.5
) +
# Adding median line with annotation
geom_vline(aes(xintercept = med_hs,
),
color = "steelblue",
linetype = "dotted",
linewidth = .8,
hjust = 0.6,
size = 3) +
annotate(
"text",
x = med_hs+0.4,
y = 0.45,
label = paste("Median: ", round(med_hs, 2)),
color = "steelblue",
size = 3.5
) +
# Adding titles, subtitles, and axes labels
labs(
title = "Uneven Distribution of Happiness Scores",
subtitle = "Median and Mean happiness indicators are similar, with various humps noted",
x = "Happiness Score"
) +
theme(
panel.grid.major = element_blank(),
plot.title = element_text(face="bold"),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
plot.background = element_rect(fill="#f5f5f5",colour="#f5f5f5")
)
Dependent Variables
Categorical
ggplot(data = wh,
aes(x = fct_rev(fct_infreq(Region)))) +
geom_bar()+
ylim(0, 45) +
geom_text(stat="count",
aes(label=paste0(after_stat(count), ", ",
round(after_stat(count)/sum(after_stat(count))*100, 1), "%")),
vjust= 0.5,
hjust = -0.2,
size= 3.5) +
labs(x = "Region",
y = "No. of Countries",
title = "Number of Countries by Region") +
theme_grey() +
theme(
plot.title = element_text(face="bold"),
axis.title.y = element_text(vjust=1, angle=0),
axis.title.x = element_text(),
plot.background=element_rect(fill="#f5f5f5",colour="#f5f5f5")
) +
coord_flip()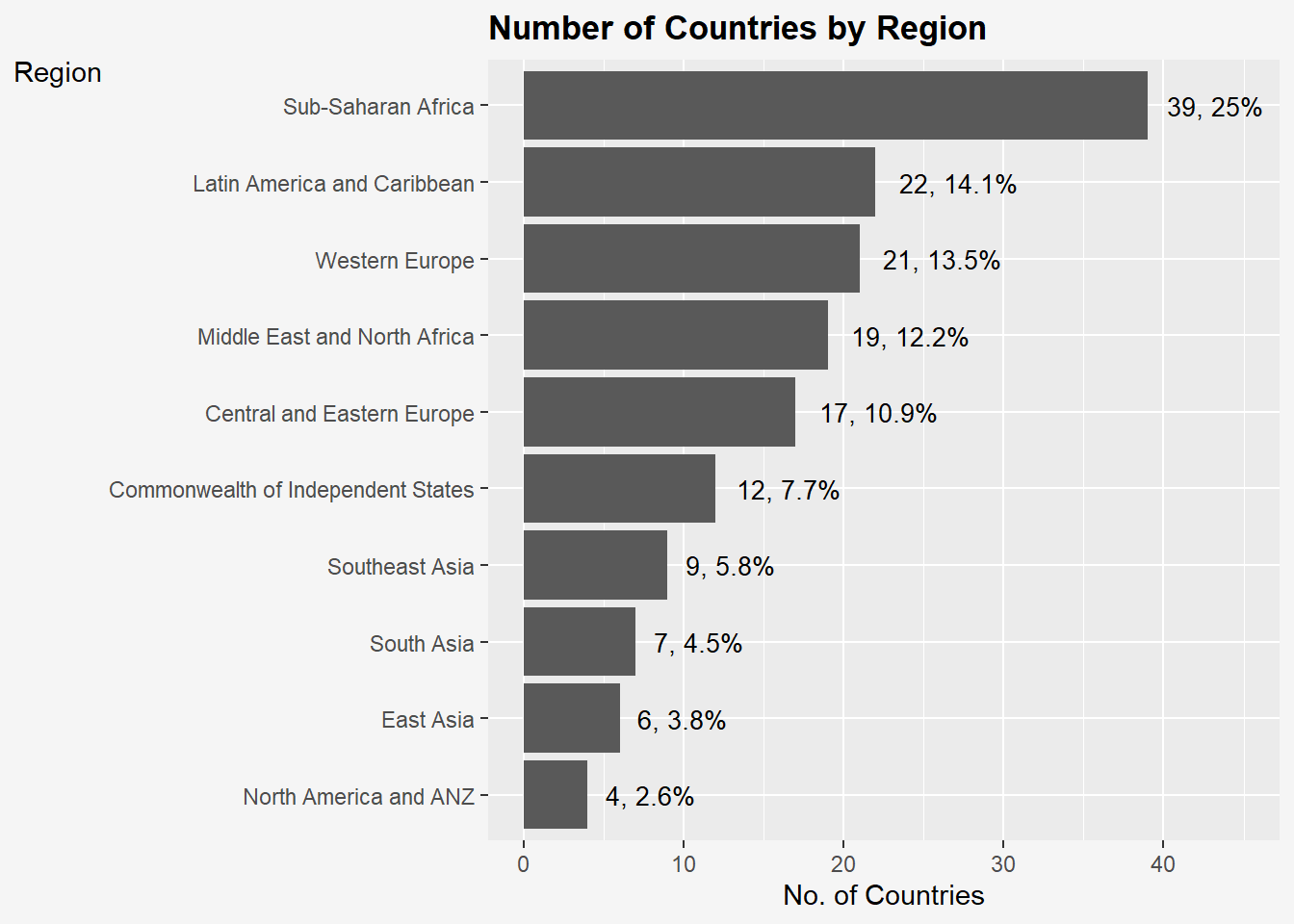
Numerical
Show the code
# Whisker-high
p_wh <-
ggplot(
wh,
aes(x = `Whisker-high`)
) +
# Density plot of target variable
geom_density(
color = "#EABEC3",
fill = "#EABEC3",
# Adjust bandwidth to reduce smoothness of density plot to reveal variations
adjust = .2,
alpha = .6
) +
# Adding titles, subtitles, and axes labels
labs(
x = "Whisker-High"
) +
theme(
panel.grid.major = element_blank(),
plot.title = element_text(face="bold"),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.x = element_text(face="bold"),
plot.background = element_rect(fill="#f5f5f5",colour="#f5f5f5")
)
p_wl <-
ggplot(
wh,
aes(x = `Whisker-low`)
) +
# Density plot of target variable
geom_density(
color = "#EABEC3",
fill = "#EABEC3",
# Adjust bandwidth to reduce smoothness of density plot to reveal variations
adjust = .2,
alpha = .6
) +
# Adding titles, subtitles, and axes labels
labs(
x = "Whisker-Low"
) +
theme(
panel.grid.major = element_blank(),
plot.title = element_text(face="bold"),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.x = element_text(face="bold"),
plot.background = element_rect(fill="#f5f5f5",colour="#f5f5f5")
)
p_dys <-
ggplot(
wh,
aes(x = Dystopia)
) +
# Density plot of target variable
geom_density(
color = "#EABEC3",
fill = "#EABEC3",
# Adjust bandwidth to reduce smoothness of density plot to reveal variations
adjust = .2,
alpha = .6
) +
# Adding titles, subtitles, and axes labels
labs(
x = "Dystopia"
) +
theme(
panel.grid.major = element_blank(),
plot.title = element_text(face="bold"),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.x = element_text(face="bold"),
plot.background = element_rect(fill="#f5f5f5",colour="#f5f5f5")
)
p_gdppc <-
ggplot(
wh,
aes(x = `GDP per capita`)
) +
# Density plot of target variable
geom_density(
color = "#EABEC3",
fill = "#EABEC3",
# Adjust bandwidth to reduce smoothness of density plot to reveal variations
adjust = .2,
alpha = .6
) +
# Adding titles, subtitles, and axes labels
labs(
x = "GDP Per Capita"
) +
theme(
panel.grid.major = element_blank(),
plot.title = element_text(face="bold"),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.x = element_text(face="bold"),
plot.background = element_rect(fill="#f5f5f5",colour="#f5f5f5")
)
p_social <-
ggplot(
wh,
aes(x = `Social support`)
) +
# Density plot of target variable
geom_density(
color = "#EABEC3",
fill = "#EABEC3",
# Adjust bandwidth to reduce smoothness of density plot to reveal variations
adjust = .2,
alpha = .6
) +
# Adding titles, subtitles, and axes labels
labs(
x = "Social support"
) +
theme(
panel.grid.major = element_blank(),
plot.title = element_text(face="bold"),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.x = element_text(face="bold"),
plot.background = element_rect(fill="#f5f5f5",colour="#f5f5f5")
)
p_lifeexp <-
ggplot(
wh,
aes(x = `Healthy life expectancy`)
) +
# Density plot of target variable
geom_density(
color = "#EABEC3",
fill = "#EABEC3",
# Adjust bandwidth to reduce smoothness of density plot to reveal variations
adjust = .2,
alpha = .6
) +
# Adding titles, subtitles, and axes labels
labs(
x = "Healthy Life Expectancy"
) +
theme(
panel.grid.major = element_blank(),
plot.title = element_text(face="bold"),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.x = element_text(face="bold"),
plot.background = element_rect(fill="#f5f5f5",colour="#f5f5f5")
)
p_freedom <-
ggplot(
wh,
aes(x = `Freedom to make life choices`)
) +
# Density plot of target variable
geom_density(
color = "#EABEC3",
fill = "#EABEC3",
# Adjust bandwidth to reduce smoothness of density plot to reveal variations
adjust = .2,
alpha = .6
) +
# Adding titles, subtitles, and axes labels
labs(
x = "Freedom to Make Life Choices"
) +
theme(
panel.grid.major = element_blank(),
plot.title = element_text(face="bold"),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.x = element_text(face="bold"),
plot.background = element_rect(fill="#f5f5f5",colour="#f5f5f5")
)
p_generosity <-
ggplot(
wh,
aes(x = Generosity)
) +
# Density plot of target variable
geom_density(
color = "#EABEC3",
fill = "#EABEC3",
# Adjust bandwidth to reduce smoothness of density plot to reveal variations
adjust = .2,
alpha = .6
) +
# Adding titles, subtitles, and axes labels
labs(
x = "Generosity"
) +
theme(
panel.grid.major = element_blank(),
plot.title = element_text(face="bold"),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.x = element_text(face="bold"),
plot.background = element_rect(fill="#f5f5f5",colour="#f5f5f5")
)
p_corruption <-
ggplot(
wh,
aes(x = `Perceptions of corruption`)
) +
# Density plot of target variable
geom_density(
color = "#EABEC3",
fill = "#EABEC3",
# Adjust bandwidth to reduce smoothness of density plot to reveal variations
adjust = .2,
alpha = .6
) +
# Adding titles, subtitles, and axes labels
labs(
x = "Perceptions of Corruption"
) +
theme(
panel.grid.major = element_blank(),
plot.title = element_text(face="bold"),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.x = element_text(face="bold"),
plot.background = element_rect(fill="#f5f5f5",colour="#f5f5f5")
)
wrap_plots(p_wh, p_wl, p_dys, p_gdppc, p_social, p_lifeexp, p_freedom, p_generosity, p_corruption) +
plot_annotation(title="Uneven Distribution across explanatory variables",
theme=theme(plot.title = element_text(hjust = 0, face ="bold"),
plot.background = element_rect(fill="#f5f5f5", colour = "#f5f5f5"),
panel.border = element_blank()))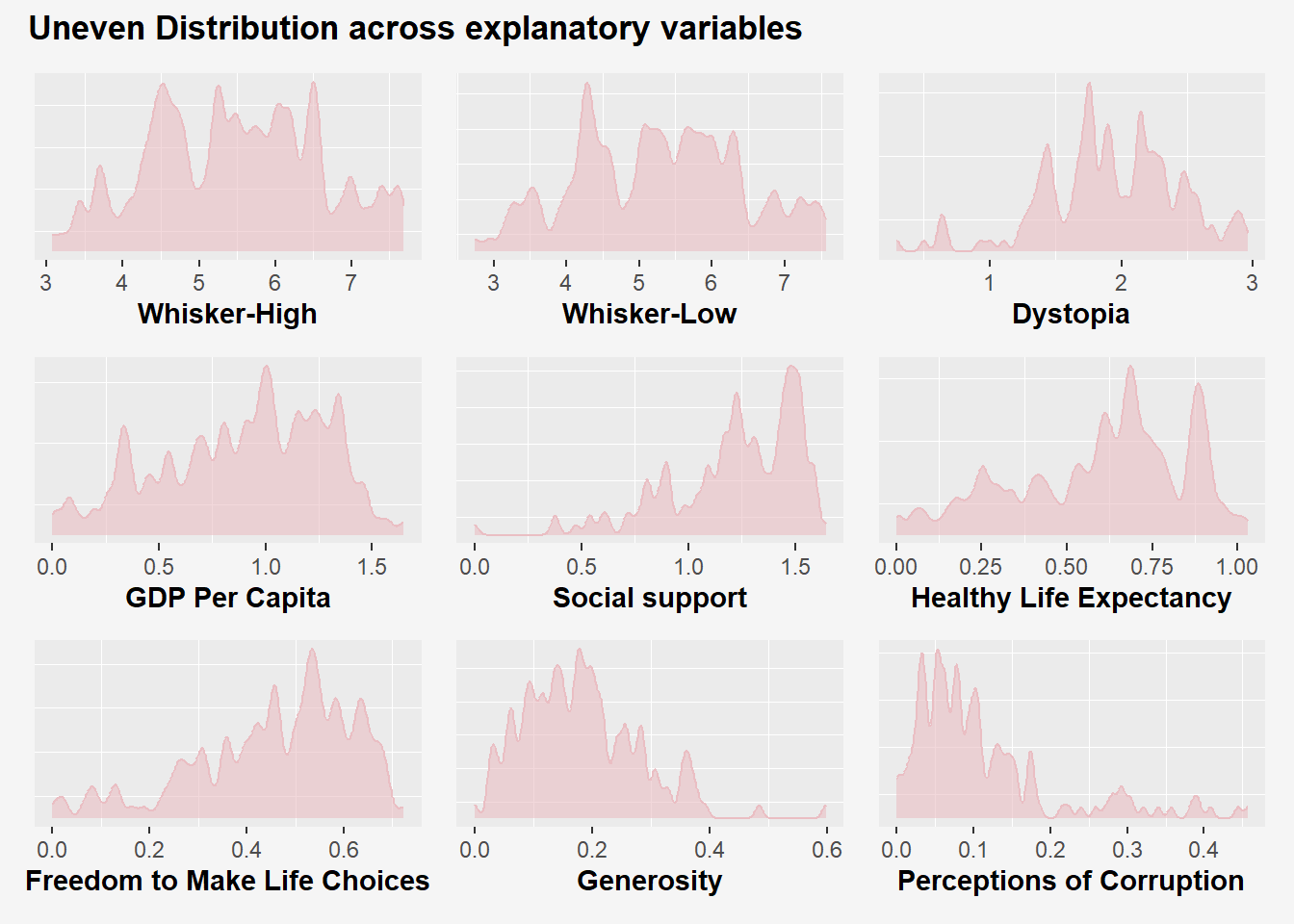
Insights
Range of each variable is different. Whisker-high/low are varies between 2 to 8, while variables like generosity and life expectancy ranges from 0 to 1.
Step 1: Changing Row Names
Next, we need to change the rows by country name instead of row number by using the code chunk below.
row.names(wh) <- wh$CountryStep 2: Transforming the data frame into a matrix
The data was loaded into a data frame, but it has to be a data matrix to make your heatmap.
The code chunk below will be used to transform wh data frame into a data matrix.
wh1 <- dplyr::select(wh, c(3, 7:12))
wh_matrix <- data.matrix(wh) #Transforms categorial variable into numerical representations.3 Static Heatmap
There are many R packages and functions can be used to drawing static heatmaps, they are:
heatmap() of R stats package for simple heatmaps.
heatmap.2() of gplots R package for enhanced heatmaps.
pheatmap() of pheatmap R package. pheatmap package also known as Pretty Heatmap. The package provides functions to draws pretty heatmaps and provides more control to change the appearance of heatmaps.
ComplexHeatmap package of R/Bioconductor package. The package draws, annotates and arranges complex heatmaps (very useful for genomic data analysis). Full reference guide of the package is available here.
superheat package: A Graphical Tool for Exploring Complex Datasets Using Heatmaps. A system for generating extendable and customizable heatmaps for exploring complex datasets, including big data and data with multiple data types. The full reference guide of the package is available here.
3.1 heatmap() of R Stats
par(bg = "#f5f5f5")
wh_heatmap <- heatmap(wh_matrix,
Rowv=NA, Colv=NA)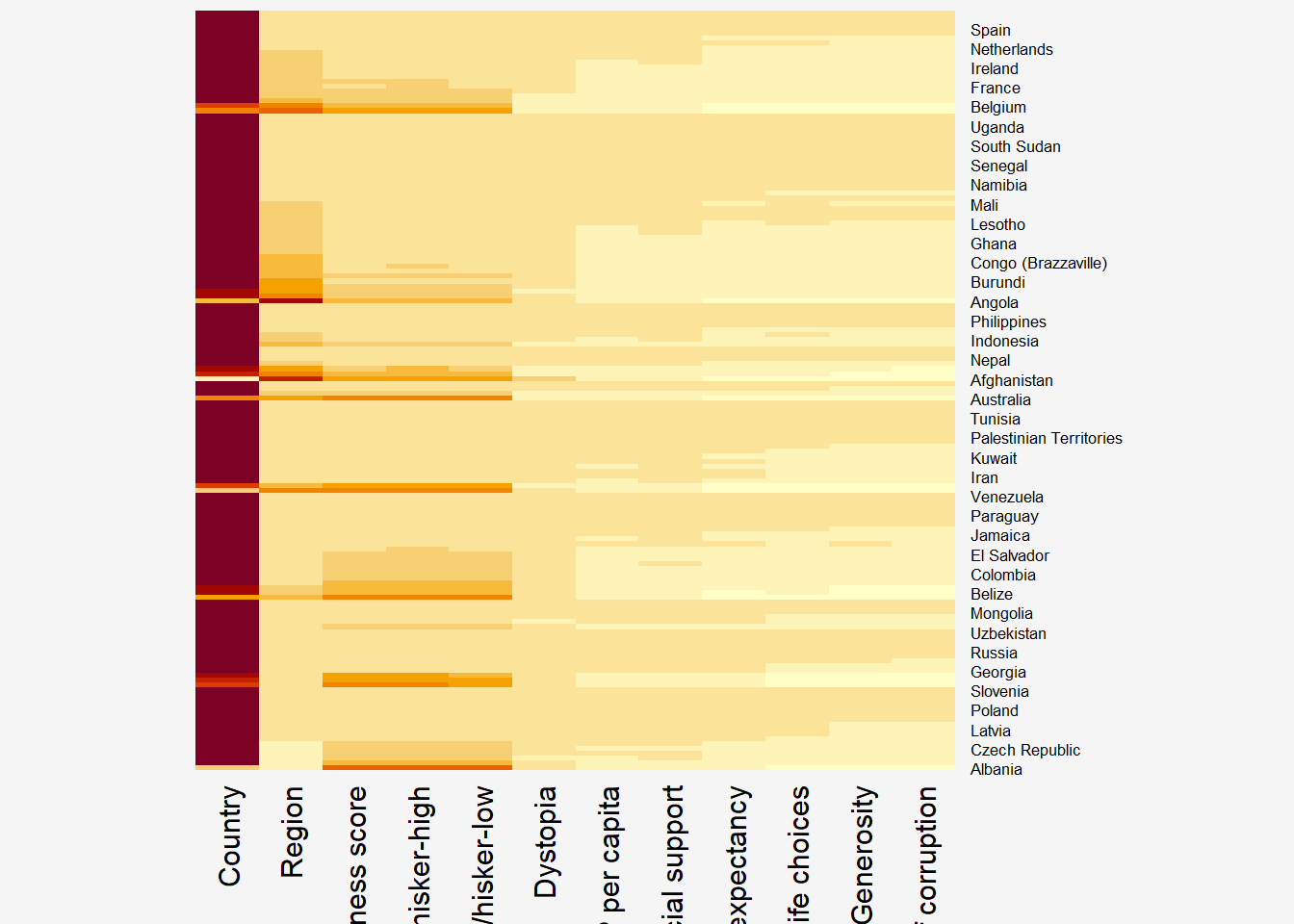
Notes from Code Chunk
By default, heatmap() plots a cluster heatmap. The arguments Rowv=NA and Colv=NA are used to switch off the option of plotting the row and column dendrograms.
par(bg = "#f5f5f5")
wh_heatmap <- heatmap(wh_matrix)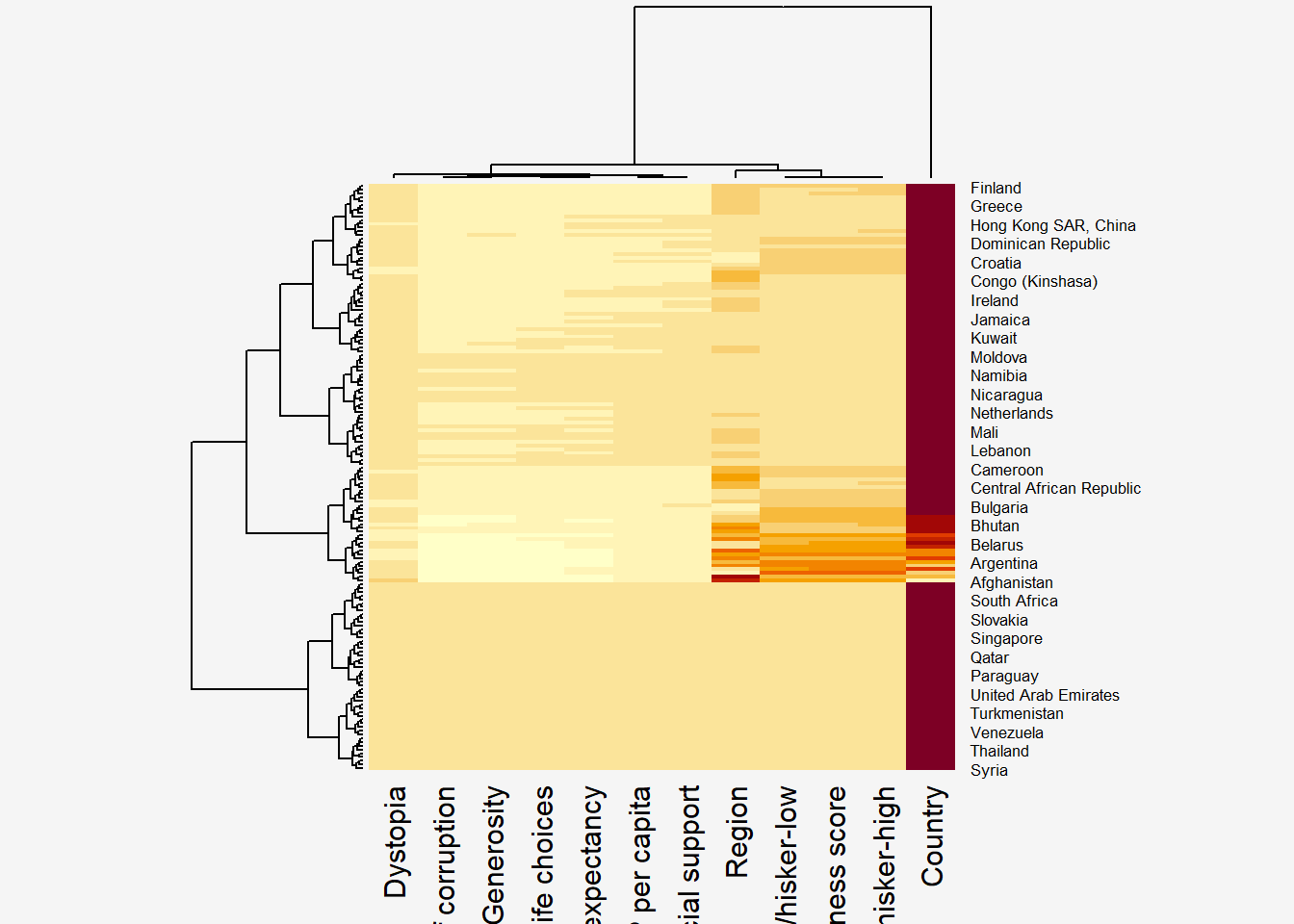
Notes from Code Chunk - The order of both rows and columns is different compare to the native wh_matrix. This is because heatmap do a reordering using clusterisation: it calculates the distance between each pair of rows and columns and try to order them by similarity. Moreover, the corresponding dendrogram are provided beside the heatmap.
Insights - This heatmap is hard to interpret since we are unable to see colours between the opposing ends of the range of the values. - Happiness Score variable have relatively higher values, which makes that the other variables with small values all look the same. Thus, we need to normalize this matrix to “scale” our data. It can be applied to rows or to columns.
par(bg = "#f5f5f5")
wh_heatmap <- heatmap(wh_matrix,
scale="column",
cexRow = 0.6,
cexCol = 0.8,
margins = c(10, 4))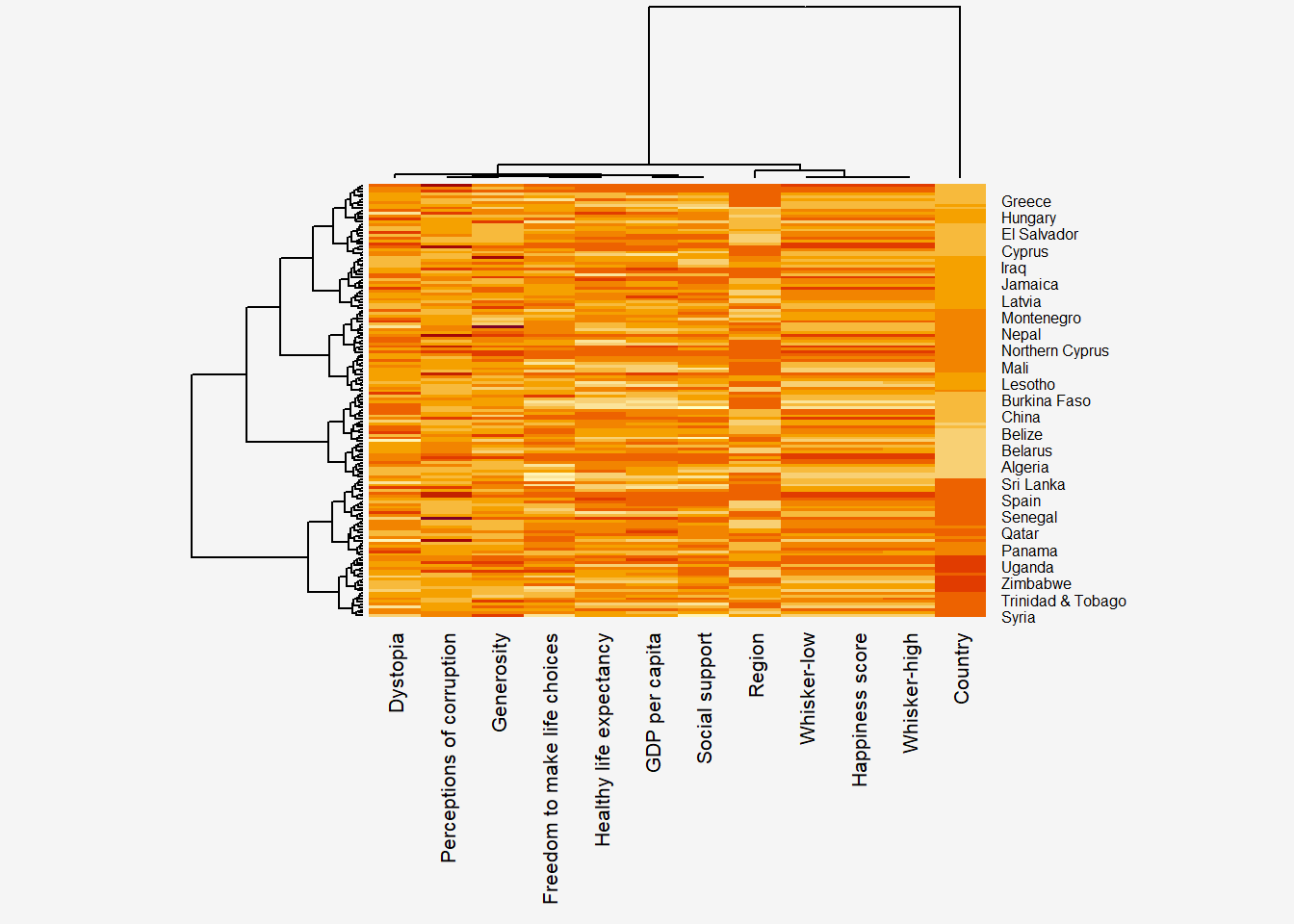
Notes from Code Chunk
- Notice that the values are scaled now.
Marginsargument is used to ensure that the entire x-axis labels are displayed completelycexRowandcexColarguments are used to define the font size used for y-axis and x-axis labels respectively.
4 Interactive Heatmap
heatmaply is an R package for building interactive cluster heatmap that can be shared online as a stand-alone HTML file. It is designed and maintained by Tal Galili.
4.1 Working with heatmaply
head(mtcars,5) mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2par(bg = "#f5f5f5")
heatmaply(mtcars)The code chunk below shows the basic syntax needed to create an interactive heatmap by using heatmaply package.
heatmaply(wh_matrix[, -c(1, 2, 4, 5)], #<<< Removes Country, Region, Whisker-High/Low
fontsize_col = 7,
fontsize_row = 4,
margins = c(60,130,10,10)) Note that:
- Different from heatmap(), for heatmaply() the default horizontal dendrogram is placed on the left hand side of the heatmap.
- The text label of each raw, on the other hand, is placed on the right hand side of the heat map.
- When the x-axis marker labels are too long, they will be rotated by 135 degree from the north.
4.2 Data transformation
When analysing multivariate data set, it is very common that the variables in the data sets includes values that reflect different types of measurement. In general, these variables’ values have their own range. In order to ensure that all the variables have comparable values, data transformation are commonly used before clustering.
Three main data transformation methods are supported by heatmaply(), namely:
scale,
normalise and
percentilise.
4.2.1 Scaling method
When all variables are came from or assumed to come from some normal distribution, then scaling (i.e.: subtract the mean and divide by the standard deviation) would bring them all close to the standard normal distribution.
In such a case, each value would reflect the distance from the mean in units of standard deviation.
The scale argument in heatmaply() supports column and row scaling.
The code chunk below is used to scale variable values column-wise.
heatmaply(wh_matrix[, -c(1, 2, 4, 5)],
fontsize_col = 7,
fontsize_row = 4,
scale = "column")4.2.2 Normalising method
When variables in the data comes from possibly different (and non-normal) distributions, the normalize function can be used to bring data to the 0 to 1 scale by subtracting the minimum and dividing by the maximum of all observations.
- This preserves the shape of each variable’s distribution while making them easily comparable on the same “scale”.
Different from Scaling, the normalise method is performed on the input data set i.e. wh_matrix as shown in the code chunk below.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
fontsize_col = 7,
fontsize_row = 4)4.2.3 Percentising method
- This is similar to ranking the variables, but instead of keeping the rank values, divide them by the maximal rank.
- This is done by using the ecdf of the variables on their own values, bringing each value to its empirical percentile.
- The benefit of the
percentizefunction is that each value has a relatively clear interpretation, it is the percent of observations that got that value or below it.
Similar to Normalize method, the Percentize method is also performed on the input data set i.e.wh_matrix as shown in the code chunk below.
heatmaply(percentize(wh_matrix[, -c(1, 2, 4, 5)]),
fontsize_col = 7,
fontsize_row = 4)4.3 Clustering algorithm
heatmaply supports a variety of hierarchical clustering algorithm. The main arguments provided are:
distfun: function used to compute the distance (dissimilarity) between both rows and columns. Defaults to dist. The options “pearson”, “spearman” and “kendall” can be used to use correlation-based clustering, which uses as.dist(1 - cor(t(x))) as the distance metric (using the specified correlation method).
hclustfun: function used to compute the hierarchical clustering when Rowv or Colv are not dendrograms. Defaults to hclust.
dist_method default is NULL, which results in “euclidean” to be used. It can accept alternative character strings indicating the method to be passed to distfun. By default distfun is “dist”” hence this can be one of “euclidean”, “maximum”, “manhattan”, “canberra”, “binary” or “minkowski”.
hclust_method default is NULL, which results in “complete” method to be used. It can accept alternative character strings indicating the method to be passed to hclustfun. By default hclustfun is hclust hence this can be one of “ward.D”, “ward.D2”, “single”, “complete”, “average” (= UPGMA), “mcquitty” (= WPGMA), “median” (= WPGMC) or “centroid” (= UPGMC).
In general, a clustering model can be calibrated either manually or statistically.
4.3.1 Manual approach
In the code chunk below, the heatmap is plotted by using hierachical clustering algorithm with “Euclidean distance” and “ward.D” method.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
dist_method = "euclidean",
hclust_method = "ward.D",
fontsize_col = 7,
fontsize_row = 4)4.3.2 Statistical approach
In order to determine the best clustering method and number of cluster the dend_expend() and find_k() functions of dendextend package will be used.
Step 1: Determine Clustering Method First, the dend_expend() will be used to determine the recommended clustering method to be used.
wh_d <- dist(normalize(wh_matrix[, -c(1, 2, 4, 5)]), method = "euclidean") #<< finding distance matrix
dend_expend(wh_d)[[3]] dist_methods hclust_methods optim
1 unknown ward.D 0.6137851
2 unknown ward.D2 0.6289186
3 unknown single 0.4774362
4 unknown complete 0.6434009
5 unknown average 0.6701688
6 unknown mcquitty 0.5020102
7 unknown median 0.5901833
8 unknown centroid 0.6338734The output table shows that “average” method should be used because it gave the highest optimum value.
Step 2: Determine Optimal # of Clusters Next, find_k() is used to determine the optimal number of cluster.
par(bg = "#f5f5f5")
wh_clust <- hclust(wh_d, method = "average")
num_k <- find_k(wh_clust)
plot(num_k)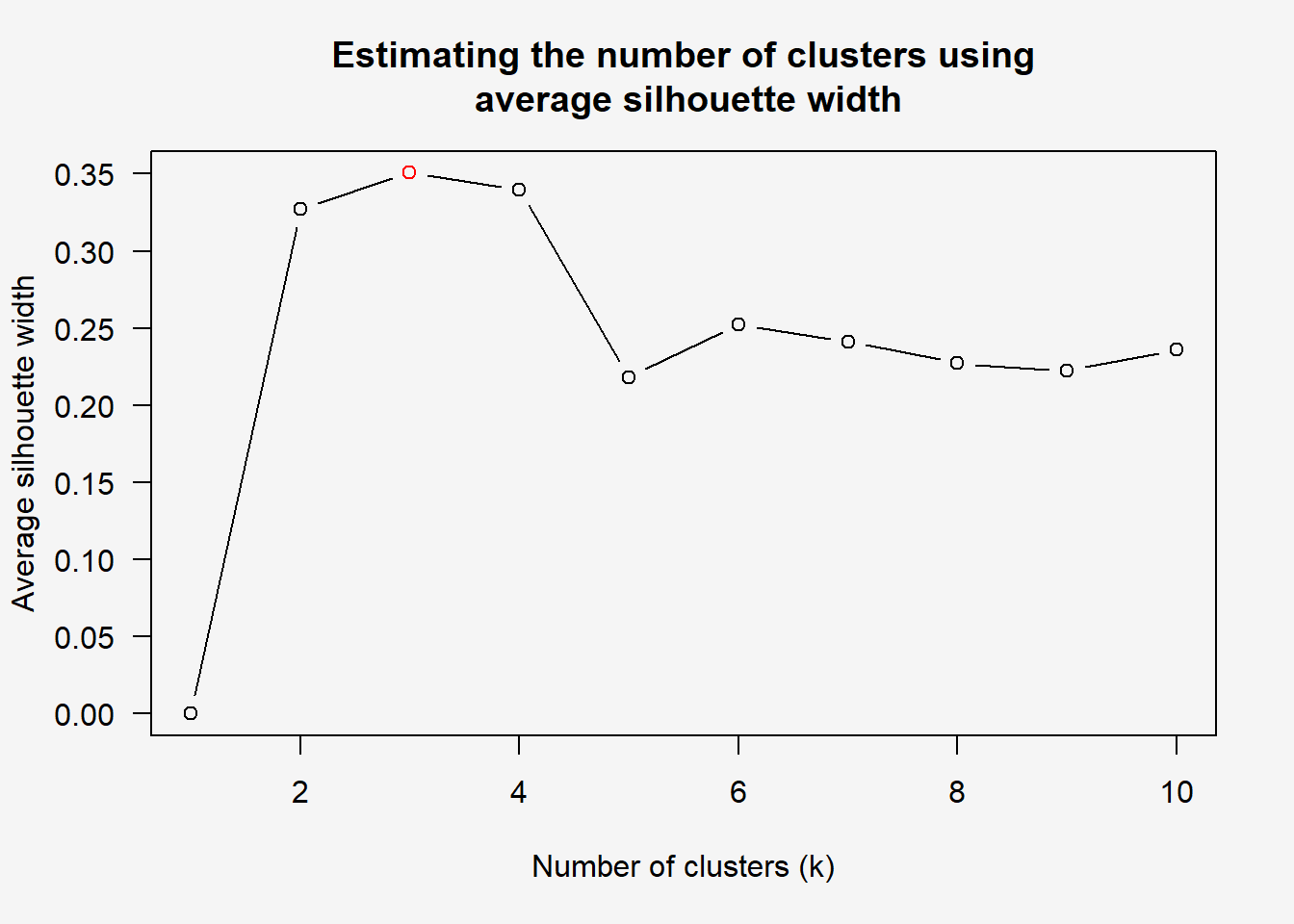
Figure above shows that k=3 would be optimal.
With reference to the statistical analysis results, we can prepare the code chunk as shown below.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
dist_method = "euclidean",
hclust_method = "average",
k_row = 3,
fontsize_col = 7,
fontsize_row = 4
)4.4 Seriation
In heatmap, every cell gets a colour, the higher the number the brighter the colour. Unfortunately, this is often a fairly unrewarding exercise, yielding graphics that look like plaid or tartan fabric. Part of the problem is that the rows and columns of a dataset often have no natural ordering, such as time, and are instead shown in alphabetical order, or else the dataset is sorted by one of the rows or columns, rather than in an order which makes patterns pop out visually.
The default options is OLO (Optimal leaf ordering) which optimizes the above criterion (in O(n^4)). Another option is GW (Gruvaeus and Wainer) which aims for the same goal but uses a potentially faster heuristic. The option mean gives the output we would get by default from heatmap functions in other packages such as gplots::heatmap.2. The option none gives us the dendrograms without any rotation that is based on the data matrix.
A less technical write-up on agglomerative clustering/seriation found here.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
seriate = "OLO",
fontsize_col = 7,
fontsize_row = 4)heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
seriate = "GW",
fontsize_col = 7,
fontsize_row = 4)The option “mean” gives the output we would get by default from heatmap functions in other packages such as gplots::heatmap.2.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
seriate = "mean",
fontsize_col = 7,
fontsize_row = 4
)heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
seriate = "none",
fontsize_col = 7,
fontsize_row = 4
)This serves as a compelling example of why it is essential to consider the capabilities and limitations of the human visual system when creating data visualizations. Simply converting numbers into graphics isn’t sufficient. Both the shuffled and organized heatmaps depicted the same data, but it’s crucial to organize the visual elements in a way that allows the human eye to perceive patterns effectively. Heatmaps, in particular, demonstrate that we can discern patterns more easily when they are presented in a spatially continuous manner. Our ability to interpret data is significantly influenced by the arrangement and proximity of the information displayed.
4.5 Working with colour palettes
The default colour palette uses by heatmaply is viridis. heatmaply users, however, can use other colour palettes in order to improve the aestheticness and visual friendliness of the heatmap.
In the code chunk below, the Blues colour palette of rColorBrewer is used.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
dist_method = "euclidean",
hclust_method = "average",
k_row = 3,
seriate = "OLO",
colors = Blues,
fontsize_col = 7,
fontsize_row = 4
)4.6 Aesthetic Changes
Beside providing a wide collection of arguments for meeting the statistical analysis needs, heatmaply also provides many plotting features to ensure cartographic quality heatmap can be produced.
In the code chunk below the following arguments are used:
k_row is used to produce 5 groups.
margins is used to change the top margin to 60 and row margin to 200.
fontsize_row and fontsize_col are used to change the font size for row and column labels to 4.
main is used to write the main title of the plot.
xlab and ylab are used to write the x-axis and y-axis labels respectively.
Explanation of margin parameter: numeric vector of length 4 (default is c(50,50,NA,0)) containing the margins (see layout) for column, row and main title names, respectively. The top margin is NA by default. If main==“” then the top margin will be set to 0, otherwise it will get 30. For a multiline title a larger default for the 3rd element should be set. The right margin is NA by default, meaning it will be zero if row_dend_left is FALSE, or 100 if row_dend_left is TRUE.
heatmaply(normalize(wh_matrix[, -c(1, 2, 4, 5)]),
Colv=NA,
seriate = "none",
colors = Blues,
k_row = 5,
margins = c(NA,200,60,NA),
fontsize_row = 4,
fontsize_col = 5,
main="World Happiness Score and Variables by Country, 2018 \nDataTransformation using Normalise Method",
xlab = "World Happiness Indicators",
ylab = "World Countries"
)5 Reference
- Kam, T.S. (2023). Visual Correlation Analysis.